|
Jiating Yu (于嘉汀) I'm currently a assistant professor at Nanjing University of Information Science and Technology in China. Before joining NUIST, I got my Ph.D. in Operations Research at Academy of Mathematics and Systems Science, Chinease Academy of Sciences , advised by Professor Ling-Yun Wu. I received my B.S. in Mathematics and Applied Mathematics at Xiangtan University in 2014, where I spent four wonderful years of my life. |

|
ResearchI'm interested in bioinformatics, computational biology, machine learning and optimization. |
|
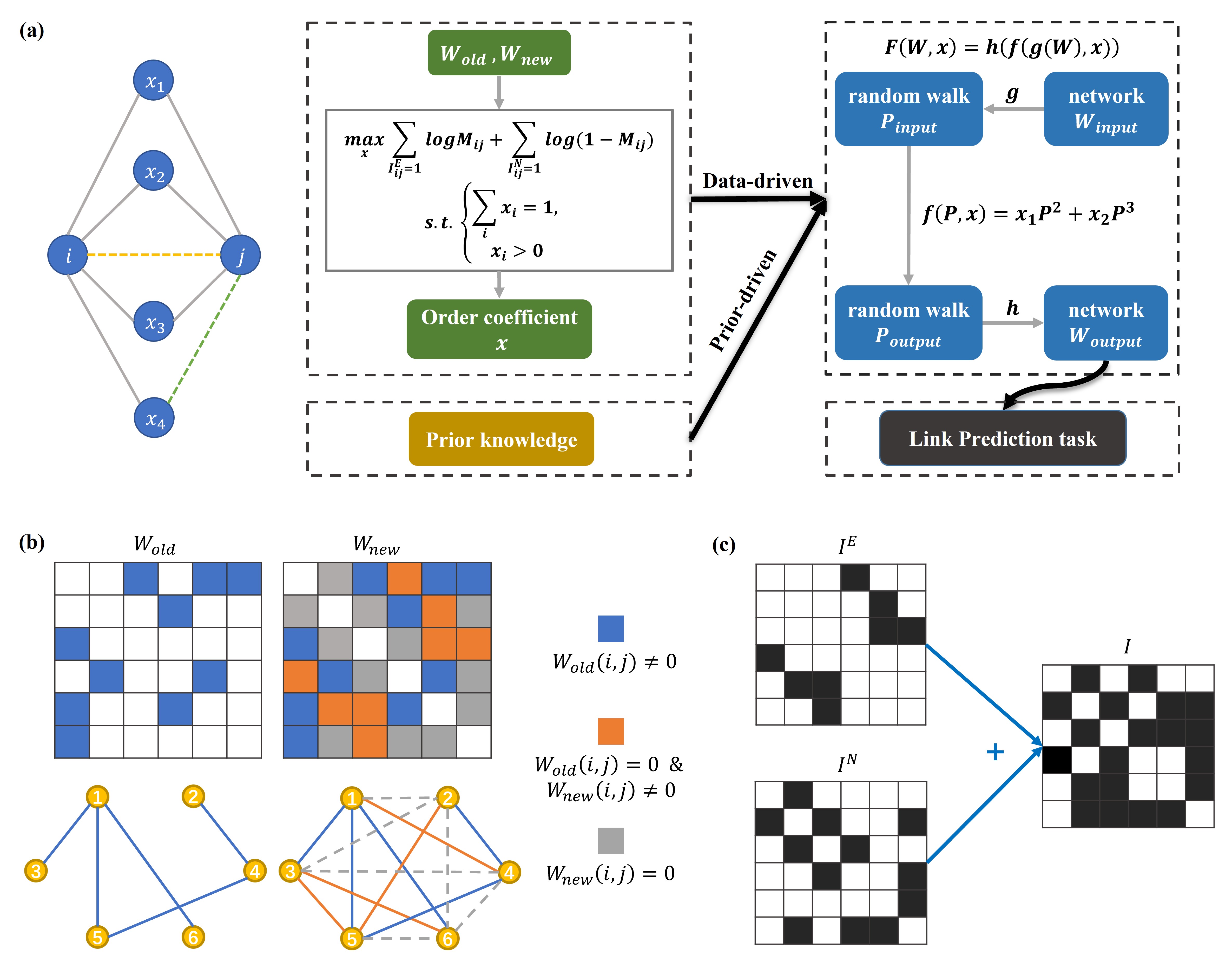
Multiple Order Local Information model for link prediction in complex networks
Jiating Yu, Ling-Yun Wu Physica A: Statistical Mechanics and its Applications, 2022 bibtex / code We proposed a novel method named Link prediction using Multiple Order Local Information (MOLI) that exploits the local information from the neighbors of different distances, with parameter that can be a prior-driven based on prior knowledge, or data-driven by solving an optimization problem on observed networks. MOLI defined a local network diffusion process via random walks on the graph, resulting in better use of network information. We show that MOLI outperforms the other 12 widely used link prediction methods on 15 different types of simulated and real-world networks. We also conclude that there are different patterns of local information utilization for different networks, including social networks, communication networks, biological networks, etc. |
|
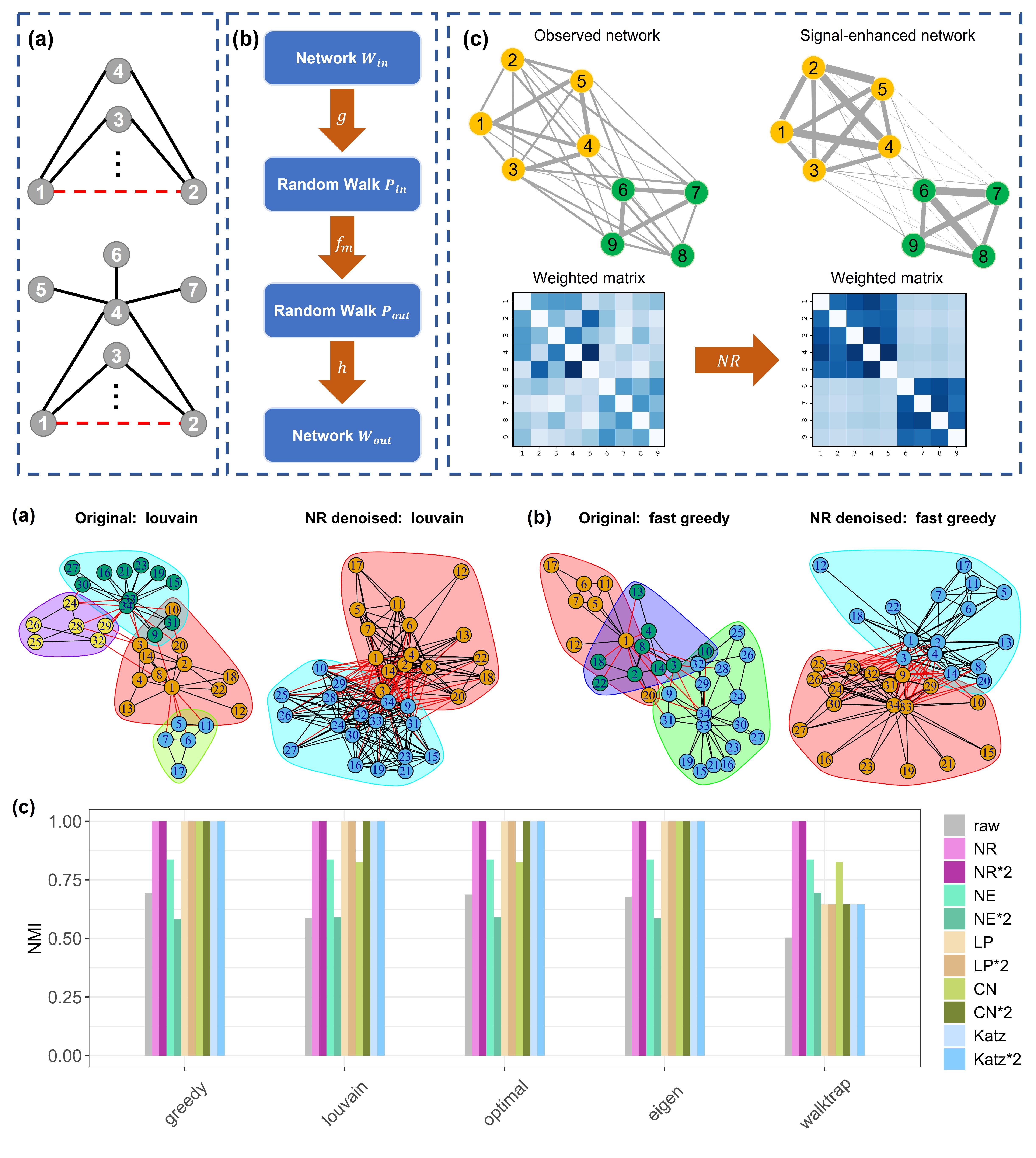
Network Refinement: Denoising complex networks for better community detection
Jiating Yu, Jiacheng Leng, Duanchen Sun, Ling-Yun Wu Physica A: Statistical Mechanics and its Applications, 2023 bibtex / code In this study, we highlighted the necessity of using network denoising as a pre-processing step to improve the performance of community detection. We proposed a novel network denoising method, called Network Refinement (NR), which used a global diffusion process defined by random walk on graph to enhance the self-organization properties of complex networks. NR took a noisy network as input and output a denoised network with clearer community structure by adjusting the edge weights. We have proved that NR can be understood as a degree normalized version of the Katz index, which renders paths with higher intermediate node degrees less important because their information is dispersed through more adjacent edges. We have showed through sufficient numerical experiments that NR significantly improves the clarity of the network’s mesoscale structure, and NR can be applied as a pre-processing step to substantially boost the performance of various community detection algorithms on both simulated networks and real-world networks. |
|
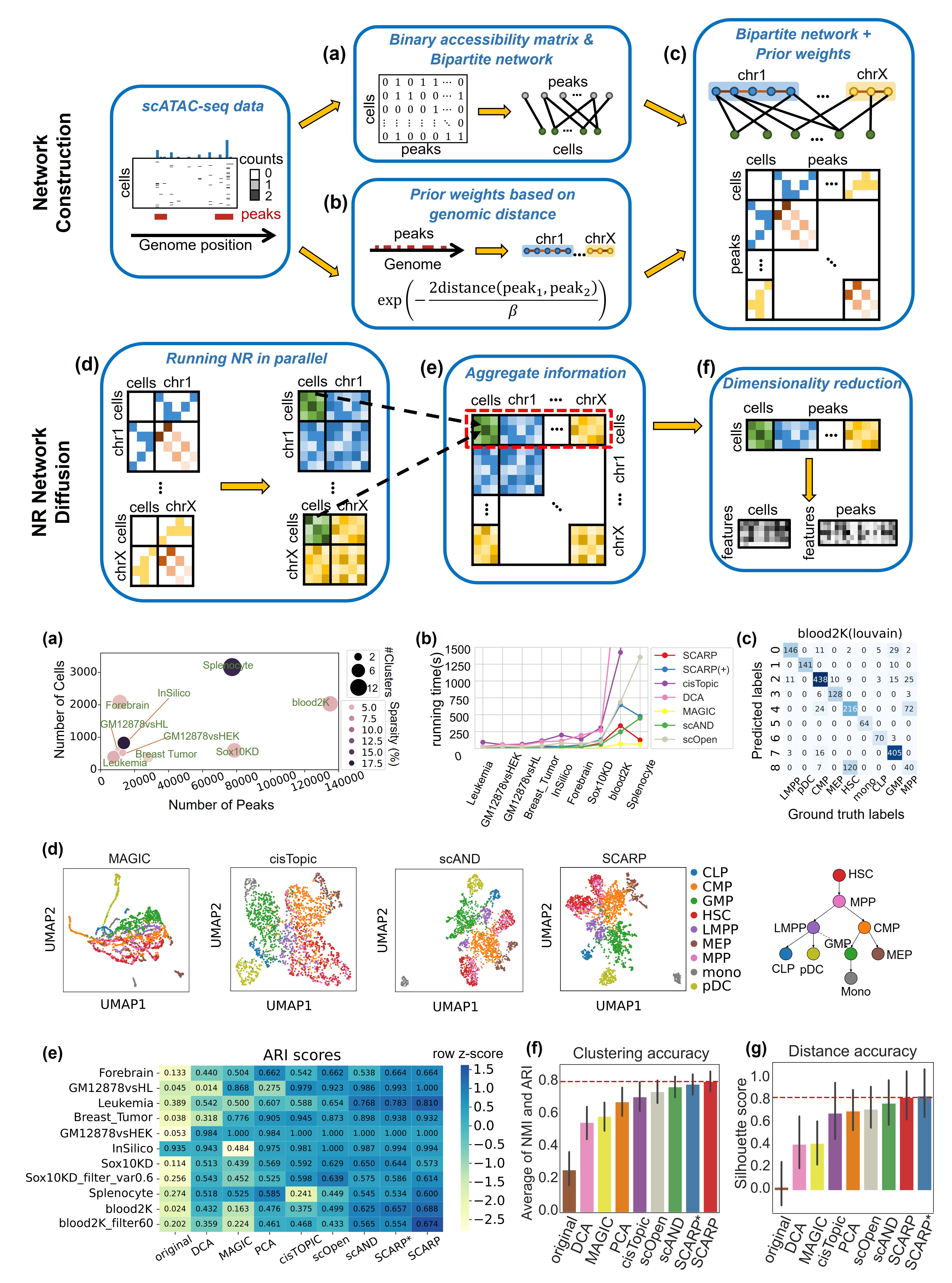
Incorporating network diffusion and peak location information for better single-cell ATAC-seq data analysis
Jiating Yu, Jiacheng Leng, Zhichao Hou, Duanchen Sun, Ling-Yun Wu Briefings in Bioinformatics, 2024 bibtex / code We proposed a novel network diffusion-based computational method to comprehensively analyze scATAC-seq data, named Single-Cell ATAC-seq Analysis via Network Refinement with Peaks Location Information (SCARP). SCARP formulates the Network Refinement diffusion method under the graph theory framework to aggregate information from different network orders, effectively compensating for missing signals in the scATAC-seq data. By incorporating distance information between adjacent peaks on the genome, SCARP also contributes to depicting the co-accessibility of peaks. These two innovations empower SCARP to obtain lower-dimensional representations for both cells and peaks more effectively. We have demonstrated through sufficient experiments that SCARP facilitated superior analyses of scATAC-seq data. |
|
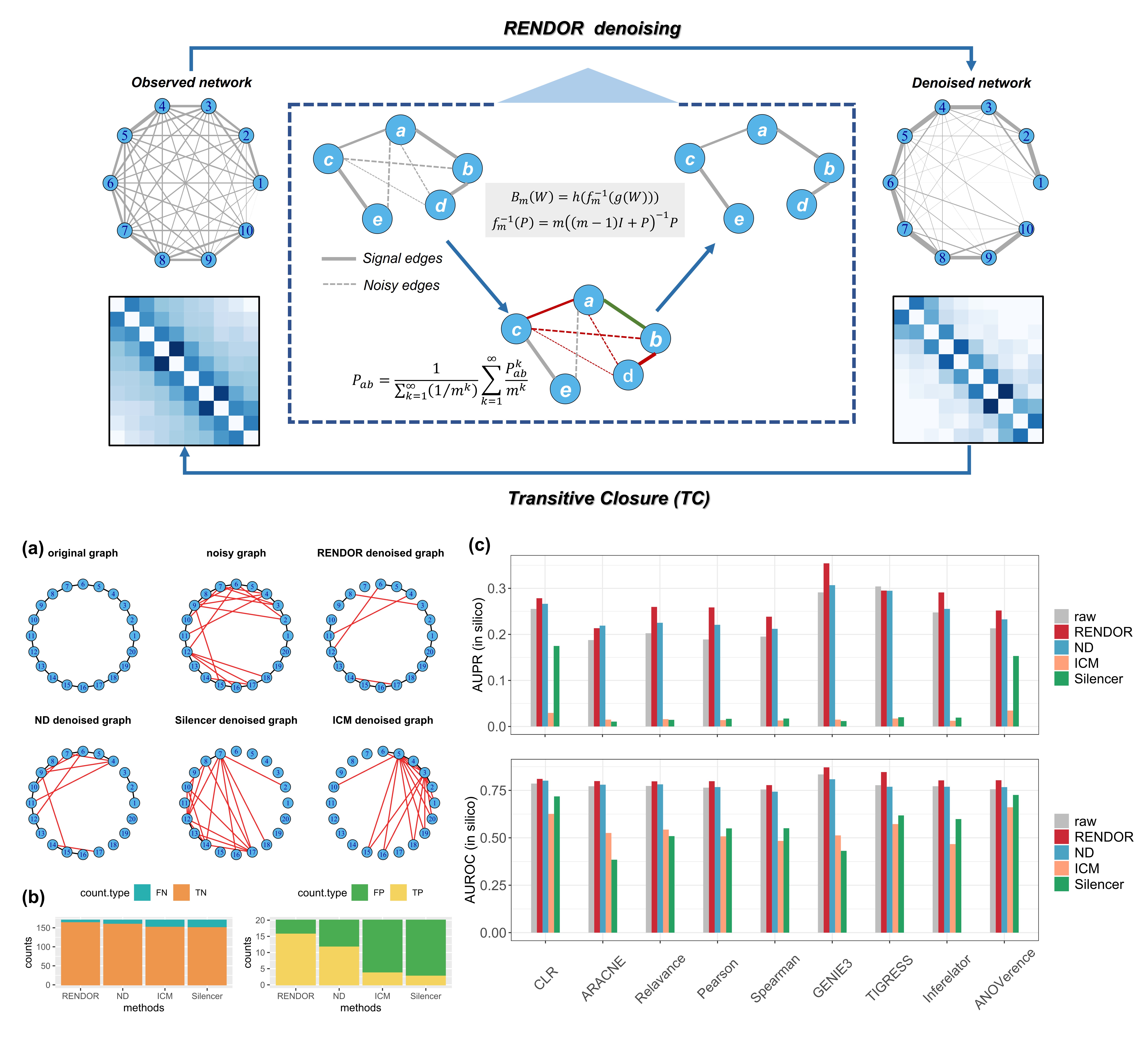
Reverse network diffusion to remove indirect noise for better inference of gene regulatory networks
Jiating Yu, Jiacheng Leng, Fan Yuan, Duanchen Sun, Ling-Yun Wu Bioinformatics, 2024 bibtex / code In this study, we proposed a novel network denoising method named REverse Network Diffusion On Random walks (RENDOR). RENDOR is designed to enhance the accuracy of GRNs afflicted by indirect effects. RENDOR takes noisy networks as input, models higher-order indirect interactions between genes by transitive closure, eliminates false-positive effects using the inverse network diffusion method, and produces refined networks as output. We conducted a comparative assessment of GRN inference accuracy before and after denoising on simulated networks and real GRNs. Our results emphasized that the network derived from RENDOR more accurately and effectively captures gene interactions. This study demonstrates the significance of removing network indirect noise and highlights the effectiveness of the proposed method in enhancing the signal-to-noise ratio of noisy networks. |
Awards |
|
(*^▽^*) |
